一.流程
1.1 分析流程
1.填写配置文件 第三列ref.fa填叶绿体/线粒体cds文件的绝对路径
cp /share/nas6/xul/program/chloroplast/RNAedit/data/config.txt ./
sample1 RNA ref.fa RNA_1.fq RNA_2.fq
sample1 DNA ref.fa DNA_1.fq DNA_2.fq
sample2 RNA ref.fa RNA_1.fq RNA_2.fq
sample2 DNA ref.fa DNA_1.fq DNA_2.fq
2.生成运行命令
perl /share/nas6/xul/program/chloroplast/RNAedit/bin/RNA_edit_pipline.pl -i config.txt
perl /share/nas6/xul/program/chloroplast/RNAedit/bin/RNA_edit_pipline.v2.pl -i config.txt
3.
sh RNA_edit/01_sh/bowtie2_build.sh && sh RNA_edit/01_sh/bowtie2_aln.sh && sh RNA_edit/01_sh/sort_sam.sh && sh RNA_edit/01_sh/get_coverage.sh && sh RNA_edit/01_sh/filter_coverage.sh && sh RNA_edit/01_sh/get_final_RNA_edit.sh
1.2 绘图程序
python /share/nas1/yuj/script/rna-editing/merge_and_output.py -i1 Tilia_mandshurica_final_rna_edit_info.txt -i2 Tilia_mandshurica_mt1_final_RNA_edit.txt -o result.txt
python /share/nas1/yuj/script/rna-editing/analyze_and_visualize.py -i result.txt -o result.png
for i in {1..3};do python /share/nas1/yuj/script/rna-editing/merge_and_output.py -i1 Pmtrep/Tilia_mandshurica_final_rna_edit_info.xls -i2 "RNA_edit/06_RNA_edit/Tilia_mandshurica_mt"$i$"_final_RNA_edit.xls" -o result.$i.txt && python /share/nas1/yuj/script/rna-editing/analyze_and_visualize.py -i result.$i.txt -o result.$i.png;done
二.结果说明
2.1 方法及参数
使用 Bowtie2 version 2.3.5.1将转录组数据比对到CDS 序列上,获取bam 文件,然后使用samtools(1.9)筛选比对质量值不小于40的序列,根据测序数据与CDS 序列的比对信息,使用脚本找到测序数据中存在的snp,保留最低深度为3的 snp 作为潜在的 RNA 编辑位点。
2.2 结果readme
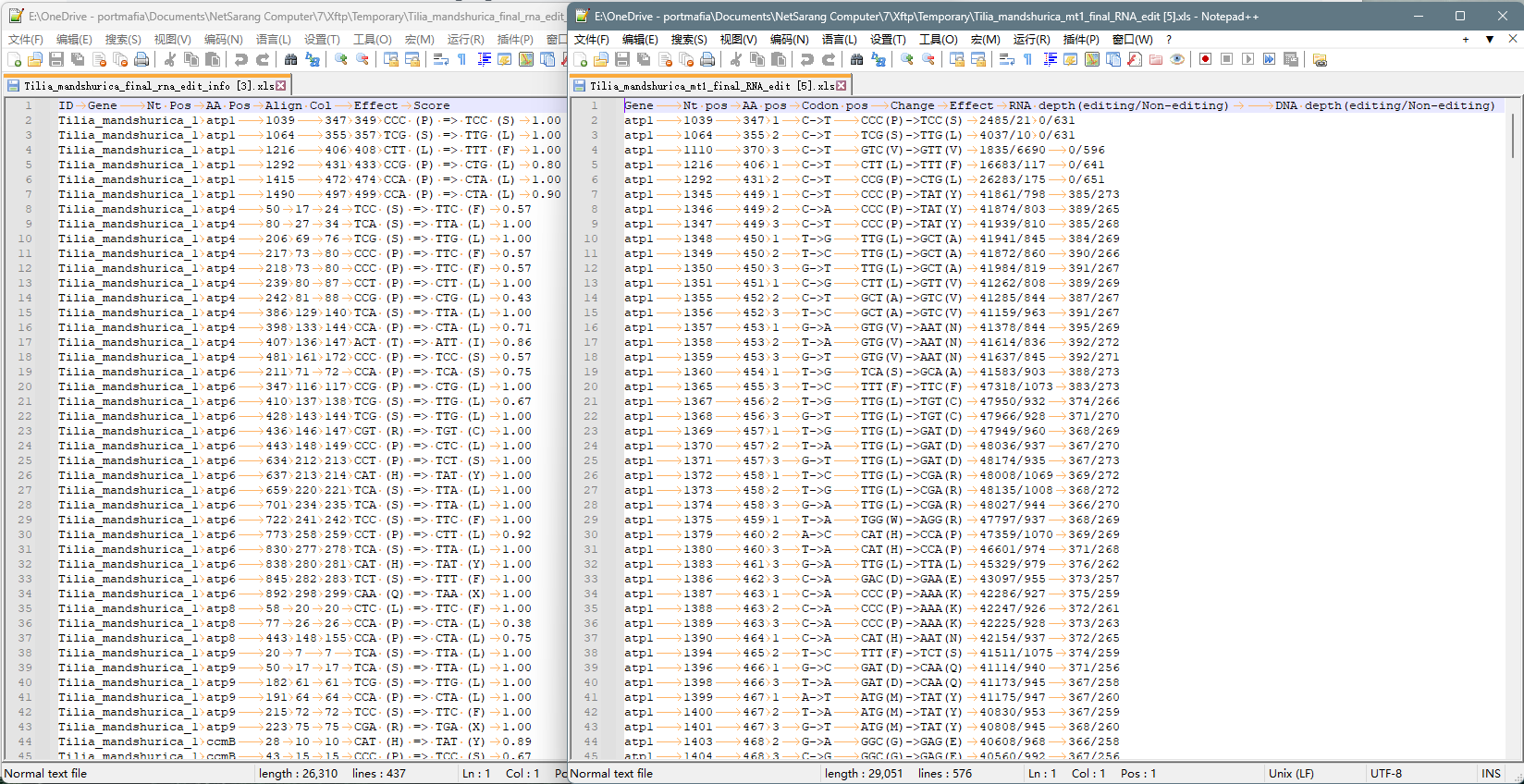
2.2.1 rna编辑预测
ID Gene Nt Pos AA Pos Align Col Effect Score
Tilia_mandshurica_1 atp1 1039 347 349 CCC (P) => TCC (S) 1.00
Tilia_mandshurica_1 atp1 1064 355 357 TCG (S) => TTG (L) 1.00
Tilia_mandshurica_1 atp1 1216 406 408 CTT (L) => TTT (F) 1.00
Tilia_mandshurica_1 atp1 1292 431 433 CCG (P) => CTG (L) 0.80
Tilia_mandshurica_1 atp1 1415 472 474 CCA (P) => CTA (L) 1.00
Tilia_mandshurica_1 atp1 1490 497 499 CCA (P) => CTA (L) 0.90
ID:序列名称;
Gene:发生RNA编辑所在的基因;
Nt pos.:RNA编辑在CDS中的位置;
AA Pos:发生RNA编辑的氨基酸位置;
Align Col:发生RNA编辑所在的对齐后的位置;
Effect:RNA编辑产生的影响;
Score:该位点的得分(0-1)。
2.2.2 转录组数据验证
Gene Nt pos AA pos Codon pos Change Effect RNA depth(editing/Non-editing) DNA depth(editing/Non-editing)
atp1 1039 347 1 C->T CCC(P)->TCC(S) 2485/21 0/631
atp1 1064 355 2 C->T TCG(S)->TTG(L) 4037/10 0/631
atp1 1110 370 3 C->T GTC(V)->GTT(V) 1835/6690 0/596
atp1 1216 406 1 C->T CTT(L)->TTT(F) 16683/117 0/641
atp1 1292 431 2 C->T CCG(P)->CTG(L) 26283/175 0/651
atp1 1345 449 1 C->T CCC(P)->TAT(Y) 41861/798 385/273
atp1 1346 449 2 C->A CCC(P)->TAT(Y) 41874/803 389/265
Gene:发生RNA编辑所在的基因;
Nt pos.:RNA编辑在CDS中的位置;
AA Pos:发生RNA编辑的氨基酸位置;
Codon pos:RNA编辑在密码子中的位置;
Change:碱基的变化;
Effect:RNA编辑产生的影响;
RNA depth(editing/Non-editing):以C->T为例,在转录组数据中,editing条reads上该位置为T,Non-editing条reads上该位置为C;
DNA depth(editing/Non-editing):以C->T为例,在基因组数据中,editing条reads上该位置为T,Non-editing条reads上该位置为C。
