04叶绿体流程2-注释
0.一些个程序 路径
命令行注释工具 https://github.com/daisieh/plann 还没测试
/share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/
/share/nas1/xul/program/chloroplast/cds_analysis/src/
1.查找单个基因/share/nas6/xul/program/chloroplast/bin/cp_annotation_one_gene_by_ref_gbk2.pl
cp_annotation_one_gene_by_ref_gbk2.pl -i1 fasta/MW148820.1.fasta -i2 ref/gbk/MH042531.1.gbk -g ndhf
2.蕨类生成gbk
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/to_genbank_for_juelei.pl
3.显示gbk有多少基因
st ref.gbk
4.查找位置
/share/nas6/xul/program/chloroplast/bin/cp_find_seq_in_fa2.pl -i /share/nas1/yuj/project/chloroplast/GP-20230717-6432_20230807/ref_ann/fasta/NC_008588.1.fasta -s TGATTATTGGTGAAAGTCAAAAAATTGGTTTCCAACCAATTAAATATCAACAAAATGTTGATGGTTCGAATACATTAATATTAG -c
79688-79771:+;
一.Reference
参考挑选标准
cp_Find_fewer_genes.pl -i xxxx.ann # 查看关键信息
rps12是3个外显子
ycf1是2个基因
rpl2可能有3段,也就是有内含子,但是有时候两段中间没有碱基,这个时候三段其实就是两段
注意参考cds trna个数 做过的陆生植物一般37 38 39trna 86 87 88cds
0.ncbi在线blast比对
1.下载参考
perl /share/nas6/xul/program/chloroplast/bin/cp_get_genbank_form_ncbi_with_ACCESSION.pl -i -o
2.挑选注释
cd ref_ann/gbk
2ann- # for i in *.gbk ;do echo $i; cp_gbk2ann.pl -i $i -o $i.ann ;done 的别名
grep rps12 *.ann # 筛选rps12有3个外显子 !!!注意rps12一般是俩
grep LOCUS * | grep 202 # 简单根据年份筛选一下
## for i in *.gbk;do st $i;done # 查看数量
for i in *.gbk;do /share/nas1/yuj/program/chloroplast/annotation/statistic.sh $i;done
# 优先87 8 37,标注了pseudo更好
csplit -z -f part_ -b "%02d.gbk" sequence.gb '/^\/\/$/' '{*}' && for file in part_*.gbk; do sed -i '/^\/\/$/d;N;/^\n$/d' "$file"; locus=$(grep -m 1 "^LOCUS" "$file" | awk '{print $2}'); mv "$file" "${locus}.gbk"; done
/share/nas6/xul/program/chloroplast/bin/cp_gbk2fasta.pl -d gbk/ -o fasta
# 读取原始文件并按“//”分隔
csplit -z -f part_ -b "%02d.gbk" sequence.gb '/^\/\/$/' '{*}'
# 删除每个新文件中的“//”分隔符行及其下面的一行空行
for file in part_*.gbk; do
# 删除“//”分隔符行及其下面的一行空行
sed -i '/^\/\/$/d;N;/^\n$/d' "$file"
# 提取登录号
locus=$(grep -m 1 "^LOCUS" "$file" | awk '{print $2}')
# 重命名文件
mv "$file" "${locus}.gbk"
done
二.注释
2.1 第一遍注释
1.配置文件,打开修改
python3 /share/nas1/yuj/script/chloroplast/get_ann_cfg.py # 在项目编号目录下直接运行
cp /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/cp.anno.config.yaml ann.cfg # 备用
2.注释
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/bin/chloroplast_annotaion.pl.v2.1.pl -d analysis/assembly/ -i ann.cfg
藻类
## 藻类植物
perl /share/nas6/xul/program/chloroplast/annotation/Annotation.pl -i1 Phaeodactylum_tricornutum_FULLCP.fsa -i2 ref_ann/gbk/MN937452.1.gbk -i3 gene_anno/gene_annotaion.info.bak -o gene_anno/ -no -orf 1>gene_anno/anno.log 2>gene_anno/anno.log2
-no 不删除中间文件
-orf 使用最全的基因列表 /share/nas6/xul/program/chloroplast/annotation/ref_normal_gene_all_other.txt
2.2 (可选)根据参考修改注释
2.2.1 cds
批量blast
## 每组cds单独blast
cp ref_ann analysis/annotation/*/gene_anno -rf
cd analysis/annotation/*/gene_anno
mkdir ann_tmp
1.提取出cds及对应蛋白序列
python3 /share/nas1/yuj/script/chloroplast/annotation/cp_from_gbk_get_cds.py -i ref_ann/gbk/ -o ref_ann/out -d
cp ref_ann/out/cds/*.fasta ann_tmp/ && python3 /share/nas1/yuj/script/chloroplast/phytree/cp_extract_and_mafft_V2.0.py -i ann_tmp/ -o1 gene/extract -o2 gene/mafft -c2
停顿几秒等比对完
cp *.fsa ann_tmp && cd ann_tmp
for i in cds*.fasta;do ir.py -s7 -n 11 -i $i -o ${i%.fasta}.pep;done
2.利用核酸和蛋白序列blast比对
for i in cds*.fasta;do blastn -query $i -subject *.fsa -outfmt 6 -max_hsps 1 > $i.log;done
for i in cds*.fasta;do tblastn -query ${i%.fasta}.pep -subject *.fsa -outfmt 6 -max_hsps 1 > ${i%.fasta}.pep.log;done
for i in cds_*.log;do echo $i;cut -f 1,7,8,9,10 $i > ${i%.log}.txt;done
3.进一步处理这些比对出的位置
## 比如这个是查找nd4基因
for i in cds*txt;do grep -i -v nd4l $i| grep -i nd4;done | cut -f 2,3,4,5 | sort -k 3
2.2.2 trna
1.
cp ref_ann/out/trna/*.fasta ann_tmp/ && cd ann_tmp
2.
for i in trna*.fasta;do blastn -task blastn-short -query $i -subject *.fsa -outfmt 6 -max_hsps 1 > $i.log;done
for i in trna_*.log;do echo $i;cut -f 1,7,8,9,10 $i > ${i%.log}.txt;done
3.
## 单独找难找的trna
name="tRNA-Ala"
for i in trna*txt;do grep -i $name $i;done | cut -f 2,3,4,5 | sort -k 3
for i in trna*fasta;do grep -A 1 -i $name $i;done > $name.fa
fl $name.fa
for i in trna*fasta;do grep -A 1 -i $name $i;done| grep -v ">"
2.2.3 rrna
1.
cp ref_ann/out/rrna/*.fasta ann_tmp && cd ann_tmp
2.
for i in rrna*.fasta;do blastn -query $i -subject *.fsa -outfmt 6 -max_hsps 1 > $i.log;done
for i in rrna_*.log;do echo $i;cut -f 1,7,8,9,10 $i > ${i%.log}.txt;done
3.
for i in rrna*txt;do grep -i 16s $i;done | cut -f 2,3,4,5 | sort -k 3
## for循环直接全部处理了
for j in {rrsA,rrlA,rrfA,rrsB,rrlB,rrfB}; do echo $j;for i in rrna*txt; do grep -i $j $i ; done | cut -f 2,3,4,5 | sort -k 3 ;done > ../allrrna.blast.txt && cd ../
1 1481 6276 7756
1 1481 6276 7756
从结果里挑选最符合的
2.2.4 单个基因
单个基因blast
## cds
gene=psbL && rm $gene.*
for i in out/cds/*;do grep -A 1 -i "\b$gene\b" $i >> $gene.all.fa ;done
ir.py -s7 -n 11 -i $gene.all.fa && blastn -query $gene.all.fa -subject *_FULLCP.fsa -outfmt 6 -max_hsps 2 >$gene.all.blast && tblastn -query $gene.pep -subject *_FULLCP.fsa -outfmt 6 -max_hsps 2 >>$gene.all.blast && cat $gene.all.blast | cut -f 7,8,9,10 | sort -k 3
## rrna
gene=rrlA && rm $gene.*
for i in out/rrna/*;do grep -A 1 -i "\b$gene\b" $i >> $gene.all.fa ;done && blastn -query $gene.all.fa -subject *_FULLCP.fsa -outfmt 6 -max_hsps 1 >$gene.all.blast && cat $gene.all.blast | cut -f 7,8,9,10 | sort -k 3
## 下面这个没必要
for i in {1..20};do if [ $(($i%2)) -eq 0 ];then sed -n ""$i"p" ndhK_all.fa > $i"_ndhK";blastn -query $i"_ndhK" -subject *.fsa -outfmt 6;fi;done # 把每个物种的基因写入单个文件,然后blast
2.3 修改注释
new gene name no in ref_all: chlB 1
new gene name no in ref_all: chlL 1
new gene name no in ref_all: chlN 1
cysA Sulfate/thiosulfate import ATP-binding protein CysA
cysT sulfate/thiosulfate import ATP-binding protein CysT
new gene name no in ref_all: g7 1
new gene name no in ref_all: tic214 1
new gene name no in ref_all: ycf12 1
注释检查
- trnH-GUG 没有内含子,去掉汇总后表格里的星号,出现该情况是因为起点原因导致此基因跨首尾多了;分号
- 注意ir区基因是否对称,如3431里面trna-ile在ira区没了,只在irb有
- ndhD、rpl2、psbL 有时候起始子不是常规起始子,转录时是把中间的c变成u/t,因此在注释结果里加标识符1以便通过流程
- ycf15应该是2个或没有
- rps19跨反向重复区很多的话,末尾有对应的假基因,这种情况下共2个,否则1个;也存在俩rps19基因的情况(如3937 exl龙血树 俩rps19完整位于反向重复区)
- rps15应该有1个,也存在俩rps15(如4086 小黑麦 俩rps15完整位于反向重复区)
- ycf1两个,一真5000一假1000
- pafII改成ycf4
- pbf1改成psbN
- pafI改成ycf3
- clpP1改成clpP
- lhbA改成psbZ
- 如果没有注释结果,看看是不是给的gbk有问题,给成了ann文件
- correct_ann.log 对ir区的矫正不一定对,通过看注释结果
- anno.log2标准含错误输出,看看有没有没找到的基因,一般没有
通用步骤
0.xul版
# 取序列看蛋白
cp_add_gene_sequence.pl -i *.fsa -p 74-1039:+
cp_add_gene_sequence.pl -s ATCGTTGGAACC
# 查找单个基因
cp_annotation_one_gene_by_ref_gbk2.pl -i1 fasta/MW148820.1.fasta -i2 ref/gbk/MH042531.1.gbk -g ndhf
blastn -query t -subject *.fsa -outfmt 6
tblastn -query t -subject *.fsa -outfmt 6
1.根据ann.log先去查看final,取序列看蛋白
cp_add.py -i *.fsa -p 134809-135306:+
cp_add_gene_sequence.pl -i *.fsa -p
2.不对的话再查看tmp查找 查看基因排列
$ grep -i 'psbI' final_gene_annotaion.info2
$ for i in `grep -i 'psbI' final_gene_annotaion.info2 | awk '{print $2}'` ;do cp_add.py -i *.fsa -p $i;done > psbI_ann.log
# $ grep 'CDS' ref_ann/gbk/登录号.gbk.ann
# $ for i in `grep 'CDS' ref_ann/gbk/登录号.gbk.ann | awk '{print $2}'` ;do mt_add.py -n 5 -i ref_ann/fasta/登录号.fasta -p $i;done > ref_ann.log
$ vim cox1
$ tblastn -query cox1 -subject *.fsa -outfmt 6
3.根据记录的笔记检查数量
提取假基因脚本或者gbk搜索pseudo
cp_get_some_gene_seq.pl -i gbk/xxx.gbk -p > pseudo.fa
blastn -query pseudo.fa -subject *.fsa -outfmt 6
4.检查结果
1)注意一些特意更改起点的项目
count=0 && for i in $(grep 'CDS' final_gene_annotaion.info2 | awk '{print $3 "_:_" $2}'); do count=$((count+1)) && echo "" && echo "###############" $count ":" $(echo "$i" | awk -F "_:_" '{print $1}') "###############" && cp_add.py -i *.fsa -p $(echo "$i" | awk -F "_:_" '{print $2}') -f;done > sample_ann.log
2)根据情况修改调整
mt_move_pos.py -i final_gene_annotaion.info2 -o final_gene_annotaion.info1 -ln 113 -n1 0 -n2 1
count=0 && for i in $(grep 'CDS' final_gene_annotaion.info1 | awk '{print $3 "_:_" $2}'); do count=$((count+1)) && echo "" && echo "###############" $count ":" $(echo "$i" | awk -F "_:_" '{print $1}') "###############" && cp_add.py -i *.fsa -p $(echo "$i" | awk -F "_:_" '{print $2}') -f;done > sample_ann.log
3)再填上假基因ycf1
4)检查
cp_change_annotation.v2.pl -i final_gene_annotaion.info1 -o final_gene_annotaion.info && cp_Find_fewer_genes.pl -i final_gene_annotaion.info
cp_change_annotation.v2.pl -i final_gene_annotaion.info2 -o final_gene_annotaion.info && cp_Find_fewer_genes.pl -i final_gene_annotaion.info
sed -i 's/rpl2\tribosomal protein L2/rpl2\tribosomal protein L2\t1/g' final_gene_annotaion.info
sed -i 's/psbL\tphotosystem II protein L/psbL\tphotosystem II protein L\t1/g' final_gene_annotaion.info
sed -i 's/ndhD\tNADH-plastoquinone oxidoreductase subunit 4/ndhD\tNADH-plastoquinone oxidoreductase subunit 4\t1/g' final_gene_annotaion.info
sed -i 's/petL\tcytochrome b6\/f complex subunit VI/petL\tcytochrome b6\/f complex subunit VI\t1/g' final_gene_annotaion.info
count=0 && for i in $(grep 'CDS' final_gene_annotaion.info | awk '{print $3 "_:_" $2}'); do count=$((count+1)) && echo "" && echo "###############" $count ":" $(echo "$i" | awk -F "_:_" '{print $1}') "###############" && cp_add.py -i *.fsa -p $(echo "$i" | awk -F "_:_" '{print $2}') -f;done > sample_ann.log
# 排序 perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/change_annotation.v2.pl -i final_gene_annotaion.info2 -o final_gene_annotaion.info
# 检查 perl /share/nas6/xul/program/chloroplast/bin/cp_Find_fewer_genes.pl -i final_gene_annotaion.info
2.3.1 样品数量很多时
tail -n 50 anno.log
grep petL -A 5 tmp_check_pos
cp_add.py -i *.fsa -p `grep cemA -A 5 tmp_check_pos | tail -n 1`
grep rrn16 -A 5 tmp_check_pos
grep rrn23 -A 5 tmp_check_pos
2.4 第二遍注释
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/bin/chloroplast_annotaion.pl.v2.1.pl -i ann.cfg -d analysis/assembly/
-asmqc_bow # 不执行asmqc中mapping这一步,使用时要确保asmqc.ok已删除
-asmqc # 不执行asmqc,asmqc.ok存在与否都一样
## 生成gbk的程序
/share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/to_genbank.pl # 默认
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/to_genbank_for_juelei.pl # 蕨类生成gbk
未解之谜
---20240605 第一行解析错误,可能是样品名中间有空格,因为通常第一行用的登录号,不会出现空格
bp DNA circular 13-Jun-2023
misc_feature 1..84350
/note="LSC"
misc_feature 84351..114079
/note="IRb"
misc_feature 114080..133964
/note="SSC"
misc_feature 133965..163693
/note="IRa"
三.整理结果
3.1 检查
mkdir fig && cp analysis/annotation/*/ogdraw/*.dpi300.circular.png analysis/annotation/*/asmqc_dir/coverage_plot/*.png analysis/annotation/*/asmqc_dir/cmp.dotplot.png fig
3.2 整理
1.二代 2+3代
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/cp_pip_dir.pl -i analysis -o complete_dir
2.仅HIFI
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/cp_pip_dir_hifi.pl -i analysis -o complete_dir
rm -rf complete_dir/03Asmqc/*/*.depth.svg
rm -rf complete_dir/03Asmqc/*/sample.coverage.svg
:set nu vim显示行数
四.生成报告
1.修改报告配置文件
cp /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/html_report/cp.report.cfg report.cfg
for i in `cat ann.cfg |grep gbk | cut -f 4`;do echo "#####################"$i && head $i;done
2.二代
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/html_report/report2xml.yelvti.pl -id complete_dir -cfg report.cfg
3.二+三代
## 先单独生成3代统计
perl /share/nas6/xul/program/mt2/tgs_reads_statistic/multi_tgs_qc.pl -i analysis/samples.reads.txt -o complete_dir/01Rawdata/
# samples.reads.txt第四列填上3代路径
## 报告
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/html_report/report2xml.yelvti_sandai.pl -id complete_dir -cfg report.cfg
4.仅HiFi数据
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/html_report/report2xml.yelvti_hifi.pl -id complete_dir -cfg report.cfg
五.补充
注释有问题的时候,可以换一个cds总数正常的参考
perl /share/nas6/xul/program/chloroplast/annotation/Annotation.pl -i1 analysis/annotation/*/gene_anno/*_FULLCP.fsa -i2 analysis/annotation/*/gene_anno/ref.gbk -i3 analysis/annotation/*/gene_anno/gene_annotaion.info.bak -o analysis/annotation/*/gene_anno -no 1> analysis/annotation/*/gene_anno/anno.log 2> analysis/annotation/*/gene_anno/anno.log2
# 会生成 _gene_annotaion.info2 _gene_annotaion.info 俩文件
5.0 ir区查找
for i in *.fasta;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/IR_SC_RegionFinder.pl -i $i -p ${i%.fasta} -o ./ & done
5.1 rscu
5.1.1 rscu出问题
有可能是基因名字有更改,product没有
5.1.2 单样品绘图(有数字折线)
cd rscu_analysis
perl /share/nas6/xul/program/chloroplast/rscu/draw.rscu.nu.pl -i rscu.stat.xls -o rscu.svg
svg2xxx -t png -dpi 600 rscu.svg && svg2xxx -t pdf -dpi 600 rscu.svg
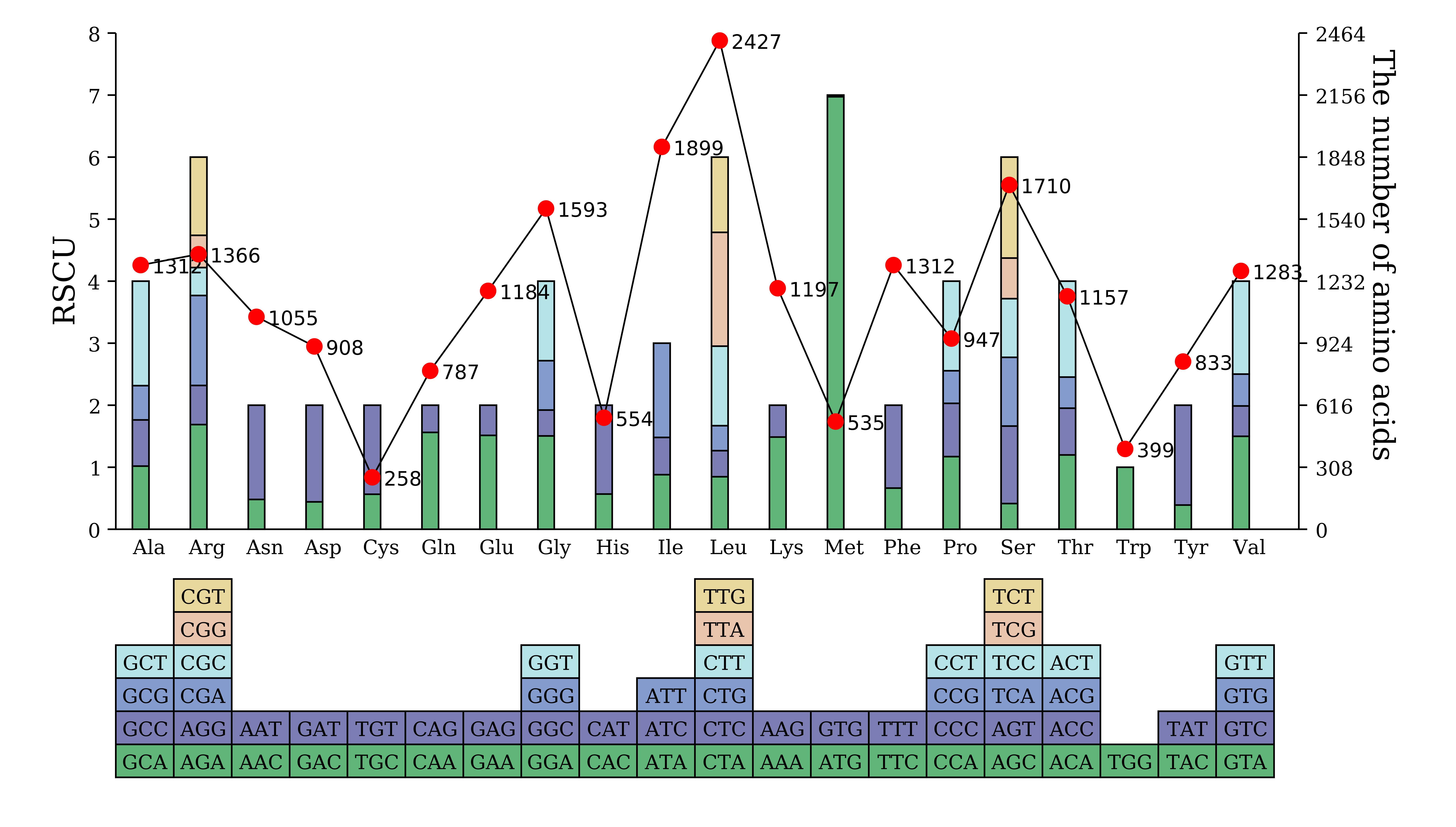
5.1.3 单样品绘图(流程图无折线)
cd rscu_analysis
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/plot_rscu_table.pl -i rscu.stat.xls -o rscu.plot.circle.svg
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation//src/plot.rscu.bar.pl -i rscu.stat.xls -o rscu.plot.bar.svg
5.1.4 多样品热图
程序路径
/share/nas1/yuj/script/chloroplast/annotation/rscu_plot/all_rscu_stat
cd ref_adv/gbk
for i in *.gbk;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/bin/../src/extrac_cds.pl -i ${i%.gbk}.gbk -o features -p ${i%.gbk} && perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/bin/../src/rscu.pl -i1 features/${i%.gbk}.cds -i2 features/${i%.gbk}.pep -o ${i%.gbk}.rscu.stat.xls ;done
python "E:\OneDrive\jshy信息部\Script\chloroplast\annotation\rscu_plot\4558_all_rscu_stat.py" ./ #当前文件夹中的rscu统计合并成一个rscu统计
tbtools导入数据,导入配置"E:\OneDrive\jshy信息部\Script\heatmap.cfg"绘图
#"E:\OneDrive\jshy信息部\Script\chloroplast\annotation\rscu_plot\heatmap.py" 热图
#"E:\OneDrive\jshy信息部\Script\chloroplast\annotation\rscu_plot\plot.py"折线图
#"E:\OneDrive\jshy信息部\Script\chloroplast\annotation\rscu_plot\plot_line.py"条形图
5.2 圈图
5.2.1 ogdraw
## 主程序
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/bin/../etc/plot_ogdraw_cpgenome_genbank/ogdraw.pl -i Medicago_sativa.gbk -p Medicago_sativa -o ogdraw
for i in `ls analysis/assembly/`;do echo $i;perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/bin/../etc/plot_ogdraw_cpgenome_genbank/ogdraw.pl -i analysis/annotation/$i/gene_anno/$i.gbk -p $i -o analysis/annotation/$i/ogdraw & done
## 圈图 png格式 dpi600
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/etc/plot_ogdraw_cpgenome_genbank/etc/GeneMap-1.1.1/bin/drawgenemap --infile gbk/MT712154.1.gbk --format png --density 600 --outfile ogdraw/test.dpi600.circular.png --useconfig /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/etc/plot_ogdraw_cpgenome_genbank/conf/OGDraw_plastid_set.xml --ir_region 85073-111624,130260-156811
5.2.2 Chloroplot
1.linux分步骤(得先手动创建ogdraw目录)
$ Rscript /share/nas1/yuj/script/chloroplast/R/Chloroplot.R $outdir/$sample/ogdraw $outdir/$sample/gene_anno/$sample.gbk $sample && convert -density 300 $outdir/$sample/ogdraw/$sample.circular.pdf $outdir/$sample/ogdraw/$sample.dpi300.circular.png
# 图片转换单拎出来
convert -density 300 *.circular.pdf $sample.dpi300.circular.png
2.windows分步骤
# 以线粒体为例,cp/mt步骤一样
# Rscript E:\OneDrive\jshy信息部\Script\chloroplast\R\Win_Chloroplot.R $outdir/$sample/ogdraw $outdir/$sample/gene_anno/$sample.gbk $sample cp/mt
$ Rscript E:\OneDrive\jshy信息部\Script\chloroplast\R\Win_Chloroplot.R F:\ F:\4939_202301\Ustilago_esculenta_MT10.gbk Ustilago_esculenta_MT10 mt
$ 复制pdf到服务器
$ convert -density 600 *.circular.pdf Ustilago_esculenta_MT10.dpi300.circular.png
3.圈图见4086项目txt(暂时不用)
圈图20220506
$ ir xxx.gbk
$ python3 /share/nas1/yuj/script/chloroplast/annotation/ogdraw.py -h
dpi300那张
4.流程里对这部分的文字描述
#-----4.3.2 叶绿体基因组图谱
&EMPTY_TAG('h3','4.3.2 叶绿体基因组图谱','','type1');
&EMPTY_TAG('p','使用Chloroplot(https://irscope.shinyapps.io/Chloroplot/)制作叶绿体基因组图谱,如下图所示:','type1');
if(glob "$indir/04Annotation/*/ogdraw/*dpi300.circular.png"){
&piclist("图$pid 叶绿体基因组图谱",'注:正向编码的基因位于圈内侧,反向编码的基因位于圈外侧。内部的灰色圈代表GC含量。',"$indir/04Annotation/*/ogdraw/*dpi300.circular.png");
}else{
&piclist("图$pid 叶绿体基因组图谱",'注:正向编码的基因位于圈内侧,反向编码的基因位于圈外侧。内部的灰色圈代表GC含量。',"$indir/04Annotation/*/ogdraw/*.circular.png");
}
5.3 asmqc
5.3.1 主程序
# 主程序
$ perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/asmqc/cpDNA_asmqc.pl -i 组装结果 -1 clean*1 -2 clean*2 -r 参考.gbk -p sample -q 物种.gbk -o asmqc_dir
---- 20231010 修改samtools路径
Bin="/share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/asmqc"
# cmd2 perl $Bin/src/gbk2fa.pl -i $refgbk -o $outdir/refseq.fa
$ perl $Bin/src/gbk2fa.pl -i $refgbk -o refseq.fa
# 生成refsseq.fa 命令3要用
# cmd3 perl $Bin/src/dotplot.pl -r $outdir/refseq.fa -q $infile -o $outdir -p cmp
$ perl $Bin/src/dotplot.pl -r refseq.fa -q $infile -o . -p cmp
# 生成共线性图
# cmd4 perl $Bin/src/annotation_cpDNA_genbank.pl -i $infile -r $refgbk -e sample -o $outdir/cmp.gbk
$ perl $Bin/src/annotation_cpDNA_genbank.pl -i $infile -r $refgbk -e sample -o cmp.gbk
# 生成cmp.gbk 和组装的gbk相比里面名称变成了sample
# cmd5 perl $Bin/src/cmp_genebank.pl -r $refgbk -q $querygbk -o $outdir/cmp.genecov_stat.xls
$ perl $Bin/src/cmp_genebank.pl -r $refgbk -q $querygbk -o cmp.genecov_stat.xls
# cmd8 perl $Bin/src/cp2circos.pl -g $outdir/cmp.gbk -b $outdir/mapping/$prefix.sort.bam -o $outdir/coverage_plot
$ perl $Bin/src/cp2circos.pl -g cmp.gbk -b mapping/*.sort.bam -o coverage_plot
# 如果只生成了mapping,运行这个程序,生成覆盖度图片 核心程序
# cmd9 perl $Bin/src/coverage_stat.pl -i $outdir -o $outdir -p $prefix
$ perl $Bin/src/coverage_stat.pl -i . -o . -p 拉丁名(同这部分主程序的$prefix)
# 生成一些统计文件 如果提示缺少文件,就是缺该组装物种的gbk
5.3.2 命令3 共线性图
cd asmqc_dir/
1.主程序
Bin="/share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/asmqc"
perl $Bin/src/dotplot.pl -r refseq.fa -q *.fsa -o . -p cmp
2.仅修改共线图颜色
sed -i '3s/repetitive$/unique/' cmp.coords.csv
Rscript /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/asmqc/src/coor_dotplot.R cmp.coords.csv cmp
## 批量替换
for i in `ls analysis/assembly/`;do echo $i;cd analysis/annotation/$i/asmqc_dir && sed -i '3s/repetitive$/unique/' cmp.coords.csv && Rscript /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/asmqc/src/coor_dotplot.R cmp.coords.csv cmp && cd - ;done
5.3.3 命令8 覆盖深度
cmd8 perl $Bin/src/cp2circos.pl -g $outdir/cmp.gbk -b
cd asmqc_dir
perl $Bin/src/cp2circos.pl -g cmp.gbk -b mapping/*.sort.bam -o coverage_plot # 会生成覆盖度的svg png
# cmp.gbk里的样品名是sample,可以把第一行相应位置修改为拉丁名 也可以使用gene_info文件夹内的gbk
FAQ:
1.图片中间物种名过长
把asmqc_dir/coverage_plot/sample.karyotype.txt中的名字改短,用下划线连接,不要有空格!
cd coverage_plot
circos="/share/nas6/zhouxy/biosoft/circos/current/bin/circos" && $circos -conf *.circos.conf && svg2xxx -t png -dpi 600 *.svg
2.图形外侧有一整圈,是因为trna跨首尾,需要修改一下
删除xxx.features.txt中最后一行或第一行
或者直接使用cmp.gbk来绘图
for i in `ls analysis/assembly/`;do echo $i;cd analysis/annotation/$i/asmqc_dir/coverage_plot && sed -i 's/Cathaya_argyrophylla_Chun_Kuang_//g' *.karyotype.txt && sed -i '$d' *.features.txt && circos="/share/nas6/zhouxy/biosoft/circos/current/bin/circos" && $circos -conf *.circos.conf && svg2xxx -t png -dpi 600 *.svg && cd - & done
分步运行
1.bowtie
cp_bowtie.align.pl -i Lanmaoa_macrocarpa_FULLMT.fsa -1 /share/nas6/xul/tmp/liulei/mogu/1444.cleaned_1.fastq.gz -2 /share/nas6/xul/tmp/liulei/mogu/1444.cleaned_2.fastq.gz -o analysis/annotation/Lanmaoa_macrocarpa/asmqc/mapping
2.筛选
samtools view -h -F 4 sample.sam > sample.map.sam
3.排序
samtools sort -@ 8 -o sample.sort.bam -T sample.sss -O bam sample.map.sam
4.深度统计
samtools depth sample.sort.bam |awk '{print "mt1",$2,$2,$3}' > sample.depth.txt # 最大值默认8000
samtools depth -m 100000 -a sample.sort.bam |awk '{print "mt1",$2,$2,$3}' > sample.depth.txt # 输出包括0的位置,最大值100000
5.找出深度最大值
awk '{max = ($NF > max) ? $NF : max} END {print max}' sample.depth.txt
6.修改配置文件
深度最大值/深度绝对路径/输出文件路径
原来文件中的长度
$ cp XXX/J5.circos.conf ./sample.circos.conf
7.画图
0)默认圈图
/share/nas6/zhouxy/biosoft/circos/current/bin/circos -conf sample.circos.conf
1)线性图1
perl /share/nas6/xul/program/chloroplast/assembly/draw_line_depth.pl *.depth.txt depth.svg && svg2xxx -t png -dpi 600 depth.svg && svg2xxx -t pdf depth.svg
# 需要手动输入显示的最大值
for i in `ls analysis/assembly/`;do echo $i-------------------------------;cd analysis/annotation/$i/asmqc_dir/coverage_plot/ && perl /share/nas1/yuj/program/chloroplast/assembly/draw_line_depth.pl *.depth.txt $i.depth.svg && svg2xxx -t png -dpi 600 $i.depth.svg && svg2xxx -t pdf $i.depth.svg && cd - & done
# 自动设置y轴最大值为最大深度+500
2)线性图2
python3 /share/nas1/yuj/script/chloroplast/annotation/draw_line_depth_v2.py *.depth.txt depth2.png
for i in `ls analysis/assembly/`;do echo $i-------------------------------;cd analysis/annotation/$i/asmqc_dir/coverage_plot/ && python3 /share/nas1/yuj/script/chloroplast/annotation/draw_line_depth_v2.py *.depth.txt $i.depth2.png && cd - & done
5.4 GC统计/基因统计
5.4.1 GC
1.有IR
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/genome.gc.stat.pl -i $genome -o $outdir/$sample/$sample.genome.gc.stat.xls
## 批量
for i in *.fasta;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/genome.gc.stat.pl -i $i -o ./${i%.fasta}.gc.stat.xls;done
2.没有IR
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/asmqc/src/genome_gc.pl -i $genome -o $outdir/$sample/$sample.genome.gc.stat.xls
5.4.2 基因统计
1.有IR
## 批量
for i in *.gbk;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/gb2tbl.pl -i $i -o ${i%.gbk}/ && perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/tbl_stat.ir.pl -i ${i%.gbk}/*.tbl -f ${i%.gbk}.fasta -o ${i%.gbk}.gene_annotation_stat.xls;done
2.没有IR
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/tbl_stat.pl -i $outdir/$sample/tbl/$sample.tbl -o $outdir/$sample/$sample.gene_annotation_stat.xls
5.5 重复序列
5.5.1 散在重复序列柱状图
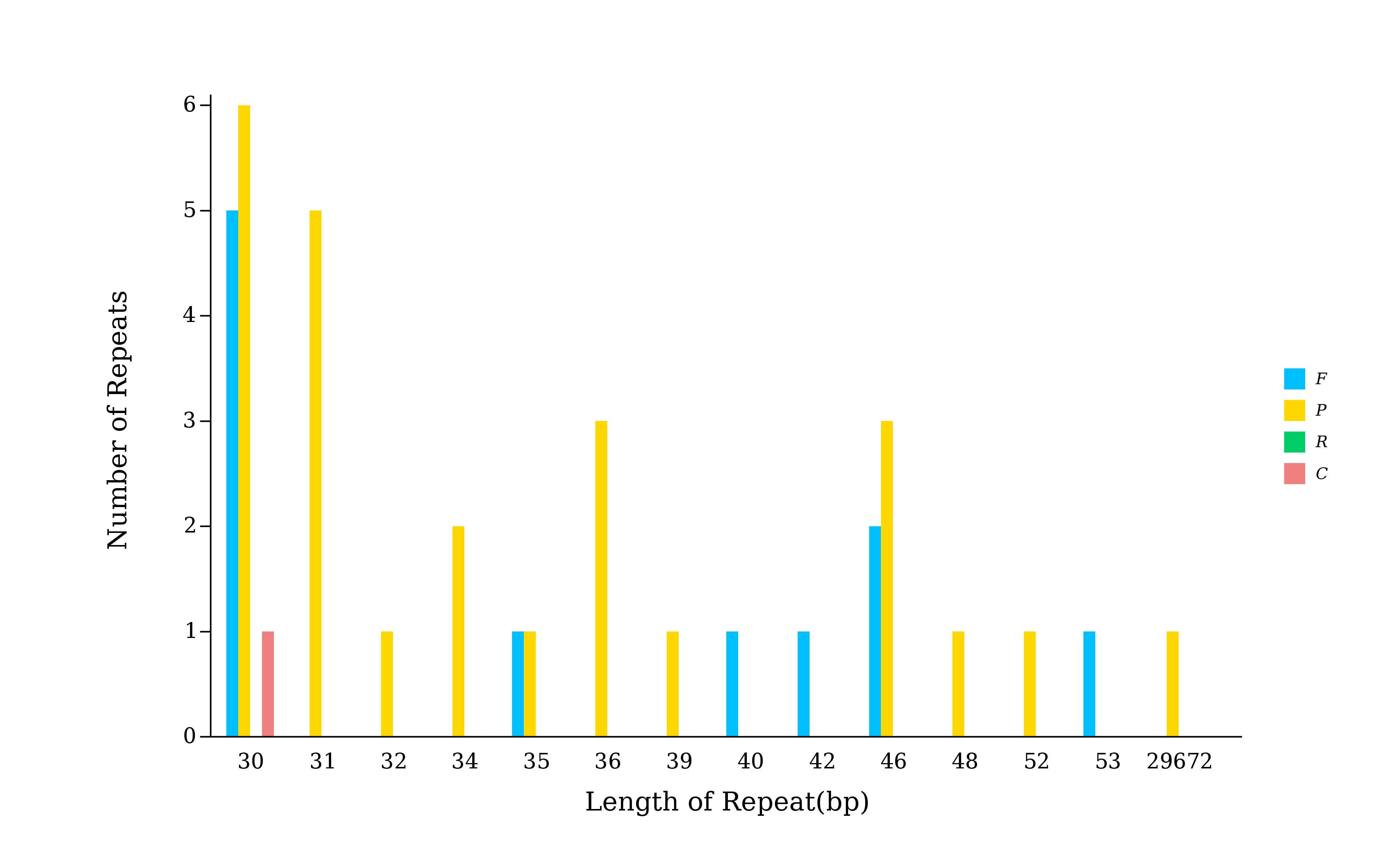
## 这一步的程序是在注释流程里该步调用的程序中出现的
## /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/reputer 程序路径
perl /share/nas6/xul/program/chloroplast/reputer/draw_reputer_len_and_tye.pl -i $outdir/$prefix.dispered.repeat.stat.xls -o $outdir/repeat.bar.svg
perl /share/nas1/yuj/program/chloroplast/reputer/draw_reputer_len_and_tye.pl -i *.dispered.repeat.stat.xls -o repeat.bar.svg # 去除图例的斜体
5.5.2 SSR 简单重复序列(流程图)
有ir:
perl $Bin/../etc/misa/src/stat.ssr.pl -i $outdir/$sample/ssr_analysis/cpSSR.misa -t $outdir/$sample/tbl/$sample.tbl -ir $ir_string -o1 $outdir/$sample/ssr_analysis/ssr.info.xls -o2 $outdir/$sample/ssr_analysis/ssr.stat.xls
perl $Bin/../etc/misa/src/plot.ssr.stat.pl -i $outdir/$sample/ssr_analysis/ssr.stat.xls -o $outdir/$sample/ssr_analysis/
perl /share/nas6/xul/program/chloroplast/ssr/draw_ssr.pl -i $outdir/$sample/ssr_analysis/number.tmp -o $outdir/$sample/ssr_analysis/ssr.number.bar
无ir:
perl $Bin/../etc/misa/src/stat.ssr.pl -i $outdir/$sample/ssr_analysis/cpSSR.misa -t $outdir/$sample/tbl/$sample.tbl -o1 $outdir/$sample/ssr_analysis/ssr.info.xls -o2 $outdir/$sample/ssr_analysis/ssr.stat.xls
perl $Bin/../etc/misa/src/plot.ssr.stat.noir.pl -i $outdir/$sample/ssr_analysis/ssr.stat.xls -o $outdir/$sample/ssr_analysis/
perl /share/nas6/xul/program/chloroplast/ssr/draw_ssr.pl -i $outdir/$sample/ssr_analysis/number.tmp -o $outdir/$sample/ssr_analysis/ssr.number.bar
5.5.3 SSR 简单重复序列(非流程图)
多样品
1.
for i in *.gbk;do mkdir ssr_analysis/${i%.gbk} -p;done
2.
for i in *.gbk;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/etc/misa/misa.pl ${i%.gbk}.fasta ssr_analysis/${i%.gbk}/cpSSR ;done
3.
for i in *.gbk;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/gb2tbl.pl -i $i -o ssr_analysis/${i%.gbk}/;done
4.
有IR:
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/etc/misa/src/stat.ssr.pl -i ssr_analysis/Lagerstroemia_caudata_NC_060355.1/cpSSR.misa -t ssr_analysis/Lagerstroemia_caudata_NC_060355.1/*.tbl -ir 84026-109650,126570-152194 -o1 ssr_analysis/Lagerstroemia_caudata_NC_060355.1/Lagerstroemia_caudata_NC_060355.1_ssr.info.xls -o2 ssr_analysis/Lagerstroemia_caudata_NC_060355.1/Lagerstroemia_caudata_NC_060355.1_ssr.stat.xls
无IR:
perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/etc/misa/src/stat.ssr.pl -i ssr_analysis/${i%.gbk}/cpSSR.misa -t ssr_analysis/${i%.gbk}/*.tbl -o1 ssr_analysis/${i%.gbk}/ssr.info.xls -o2 ssr_analysis/${i%.gbk}/ssr.stat.xls
5.生成几个物种的ssr类型统计表
cd ssr_analysis && for i in *;do cp $i/ssr.stat.xls $i.ssr.stat.xls;done && cd -
python3 /share/nas1/yuj/script/chloroplast/annotation/repeats_ssr_lsr_plot/4558_only_ssr_type_stat.py 输入目录
# cd ssr_analysis && for i in *;do cp $i/ssr.info.xls $i.ssr.info.xls;done && cd -
# "E:\OneDrive\jshy信息部\Script\chloroplast\annotation\repeats_ssr_lsr_plot\4558_only_ssr_type_stat.py"
6.excel绘制三维柱状图
单样品
只针对某个物种绘制
cp ssr.stat.xls ./
python3 /share/nas1/yuj/script/chloroplast/annotation/repeats_ssr_lsr_plot/4558_only_ssr_type_stat.py ./ # 获得only_ssr_type_stat.txt
python3 /share/nas1/yuj/script/chloroplast/annotation/repeats_ssr_lsr_plot/ssr_plot.py -i only_ssr_type_stat.txt -o ssr.bar.png
5.5.4 LSR 长散在重复序列
1.生成dispered.repeat.stat.xls
for i in *.fasta;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/reputer/find.dispered.repeat.pl -i $i -p ${i%.fassta} -o reputer;done
2.整理到一块
3.绘图
E:\OneDrive\jshy信息部\Script\chloroplast\annotation\repeats_ssr_lsr_plot\lsr_plot.py
5.5.5 合并信息绘图
需要前置文件$sample.dispered.repeat.region.xls
把散在重复文件下的分类统计表以区域拆分
"E:\OneDrive\jshy信息部\Script\chloroplast\annotation\repeats_ssr_lsr_plot\4558_repeat_region_stat.py"
生成ssr lsr柱状堆叠统计图片
"E:\OneDrive\jshy信息部\Script\chloroplast\annotation\repeats_ssr_lsr_plot\4558_all_repeat_plot.py"
5.5.6 重复序列圈图(线粒体流程)
/share/nas6/xul/program/mt2/annotation_for_multi_seq/pipline/repeat/mt2_repeat.pl 原版 线粒体参数
cd analysis/annotation/样品
mkdir repeat_circos && cd repeat_circos
perl /share/nas1/yuj/program/mt2/annotation_for_multi_seq/pipline/repeat/mt2_repeat.pl -mt *.gbk -o ./ # 与叶绿体ssr参数一致
## 仅绘图
circos="/share/nas6/xul/soft/circos/circos-0.69-6/bin/circos"
$circos -conf circos.conf
svg2xxx -t pdf repeat.svg
5.6 RSCU_PLOT
1.生成rscu表格
mkdir rscu_analysis && for i in *.gbk;do perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/extrac_cds.pl -i $i -o features -p ${i%.gbk} && touch rscu_analysis/${i%.gbk}.rscu.stat.xls && perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/annotation/src/rscu.pl -i1 features/${i%.gbk}.cds -i2 features/${i%.gbk}.pep -o rscu_analysis/${i%.gbk}.rscu.stat.xls;done
2.合并xls
# "E:\OneDrive\jshy信息部\Script\chloroplast\annotation\repeats_ssr_lsr_plot\4558_rscu_stat..py"
python3 /share/nas1/yuj/script/chloroplast/annotation/rscu_plot/4558_all_rscu_stat.py rscu_analysis
示例:
codon type Ormosia_henryi_OP450827.1 Ormosia_hosiei_OP450825.1 Ormosia_semicastrata_OP450824.1 Ormosia_xylocarpa_OP450826.1
UAA 1.7628 1.7628 1.8246 1.7628
UAG 0.6495 0.6495 0.6495 0.6495
3.绘图
tbtools绘制热图
六.批量操作
# 批量显示测序数据量
for i in $( ls /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/assembly/) ;do echo $i; ll /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/assembly/$i/pseudo/$i/1_Trimmed_Reads/$i.trimmed_P1.fq && ll /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/assembly/$i/pseudo/$i/1_Trimmed_Reads/$i.trimmed_P2.fq ;done
# 批量显示比对结果
for i in $( ls /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/assembly/) ;do echo $i; blastn -query /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/ref_ass/all_ref.fa -subject /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/assembly/$i/pseudo/$i/1_Trimmed_Reads/uni/assembly.fasta -outfmt 6 ;done
# 测序数据质控
perl /share/nas6/pub/pipline/genome-assembly-seq/mitochondrial-genome-seq/v2.0/annotation/src/fastq_qc.pl -i analysis/samples.reads.txt -o complete_dir/01Rawdata
# 批量质控 线粒体
for i in $( ls /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/assembly/) ;do echo $i; perl /share/nas6/pub/pipline/genome-assembly-seq/mitochondrial-genome-seq/v2.0/asmqc/mtDNA_asmqc.pl -i /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/ref_ass/fasta/HQ857211.1.fasta -1 /share/nas1/seqloader/xianliti/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/2.clean_data/$i/$i_*_R1_001.fastq.gz -2 /share/nas1/seqloader/xianliti/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/2.clean_data/$i/$i_*_R2_001.fastq.gz -p Gallus_gallus -q /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/ref_ass/gbk/HQ857211.1.gbk -o /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/annotation/$i/asmqc -g 2 -r /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/ref_ass/gbk/MT800385.1.gbk;done
# 自动换行查看
perl /share/nas6/pub/pipline/genome-assembly-seq/mitochondrial-genome-seq/v2.0/asmqc/mtDNA_asmqc.pl -i ref_ass/fasta/HQ857211.1.fasta -1 /share/nas1/seqloader/xianliti/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/2.clean_data/$i/$i_*_R1_001.fastq.gz -2 /share/nas1/seqloader/xianliti/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/2.clean_data/$i/$i_*_R2_001.fastq.gz -p Gallus_gallus -q /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/ref_ass/gbk/HQ857211.1.gbk -o /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/analysis/annotation/$i/asmqc -g 2 -r /share/nas1/yuj/project/20211229/GP-20211206-3811-1_longyanxueyuan_9samples_dongwu_xianliti/ref_ass/gbk/MT800385.1.gbk
# 批量质控 叶绿体
for i in $(ls /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/ref_ass/fasta);do echo $i;perl /share/nas6/pub/pipline/genome-assembly-seq/chloroplast-genome-seq/v1.2/asmqc/cpDNA_asmqc.pl -i /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/ref_ass/fasta/$i -1 /share/nas1/seqloader/yelvti/GP-20210924-3463-1_qinghaiminda_zhoulaosh_5samples_yelvti/data/2.clean_data/1/1_S0_L000_R1_000.fastq.gz -2 /share/nas1/seqloader/yelvti/GP-20210924-3463-1_qinghaiminda_zhoulaosh_5samples_yelvti/data/2.clean_data/1/1_S0_L000_R2_000.fastq.gz -p ${i%.1.fasta} -q /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/ref_ass/gbk/${i%.fasta}.gbk -o /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/${i%.1.fasta}asmqc -r /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/ref_ass/gbk/NC_053552.1.gbk;done
# 批量复制
for i in $(ls /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/ref_ass/fasta);do echo $i;cp /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/${i%.1.fasta}asmqc/coverage_plot/sample.png /share/nas1/yuj/project/GP-20210924-3463-1_20220413/analysis/assembly/1/pseudo/1/Final_Assembly/png/${i%.1.fasta}.png;done
七.基因分类
| 基因分类 | 基因分组 | 基因名称 | 基因数量 |
|---|---|---|---|
| 表达相关基因 | 核糖体 RNA 基因 | rrn16(×2)、23(×2)、4.5(×2)、5(×2) | 4 |
| 转运 RNA 基因 | trnA-CAU、trnA-UGC(×2)、trnC-GCA、trnD-GUC、trnE-UUC、trnF-GAA、trnG-GCC、trnG-GUG、trnH-CAU、trnI-CAU(×2)、trnI-GAU(×2)、trnK-UUU、trnL-CAA(×2)、trnL-UAG、trnL-UAA、trnM-CAU(×2)、trnN-GUU(×2)、trnP-UGG、trnQ-UUG、trnR-ACG(×2)、trnR-UCU、trnS-UGA、trnS-GCU、trnS-GGA、trnT-UGU、trnT-GGU、trnV-UAC、trnW-CCA、trnY-GUA | 29 | |
| 核糖体小亚基基因(SSU) | rps2、3、4、7(×2)、8、11、12(×2)、14、15(×2)、16、18、19 | 12 | |
| 核糖体大亚基基因(LSU) | rpl2(×2)、14、16、20、22、23(×2)、32、33、36 | 9 | |
| RNA 聚合酶亚基基因 | rpoA、B、C1、C2 | 4 | |
| 光合作用相关基因 | 光系统Ⅰ基因 | psaA、B、C、I、J | 5 |
| 光系统Ⅱ基因 | psbA、B、C、D、E、F、H、I、J、K、L、M、T、Z | 14 | |
| 细胞色素复合物基因 | petA、B、D、G、L、N | 6 | |
| ATP 合酶基因 | atpA、B、E、F、H、I | 6 | |
| 依赖 ATP 的蛋白酶单元 p 基因 | clpP | 1 | |
| 二磷酸核酮糖醛化酶大亚基基因 | rbcL | 1 | |
| NADH 脱氢酶基因 | ndhA、B(×2)、C、D、E、F、G、H、I、J、K | 11 | |
| 其他基因 | 成熟酶基因 | matK | 2 |
| 包裹膜蛋白基因 | cemA | 1 | |
| 乙酰辅酶 A 羧化酶亚基基因 | accD | 1 | |
| c 型细胞色素合成基因 | ccsA | 1 | |
| 转录起始因子基因 | infA | 1 | |
| 未知功能基因 | 保守开放阅读框 | ycf1(×2)、ycf2(×2)、ycf15(×2) | 3 |